PUBLICATIONS
1. Hong Y, Liu N*. Transposable elements in health and disease: molecular basis and clinical implications. Chinese Medical Journal (2025)
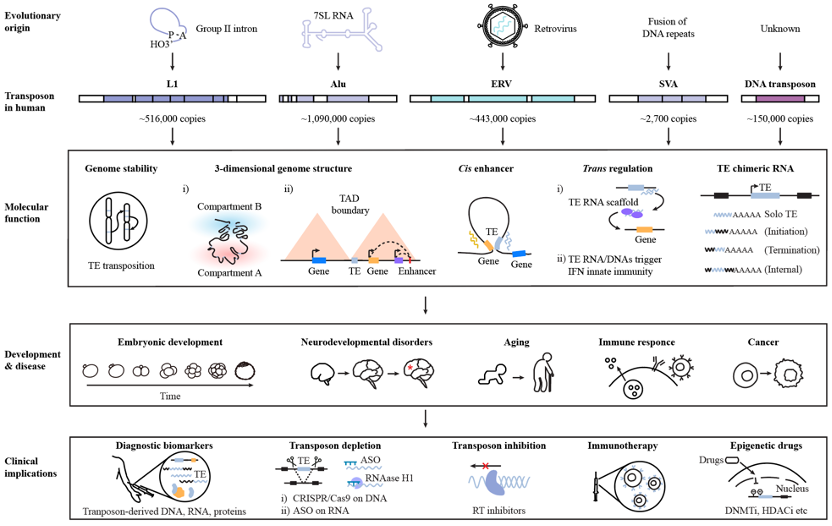
2. Zhou Z#, Zhu S#, Hong Y, Jin G, Ma R, Lin F, Zhang Y, Lee HY*, Liu N*. Composite transposons with bivalent histone marks function as RNA-dependent enhancers in cell fate regulation. Cell (2025).
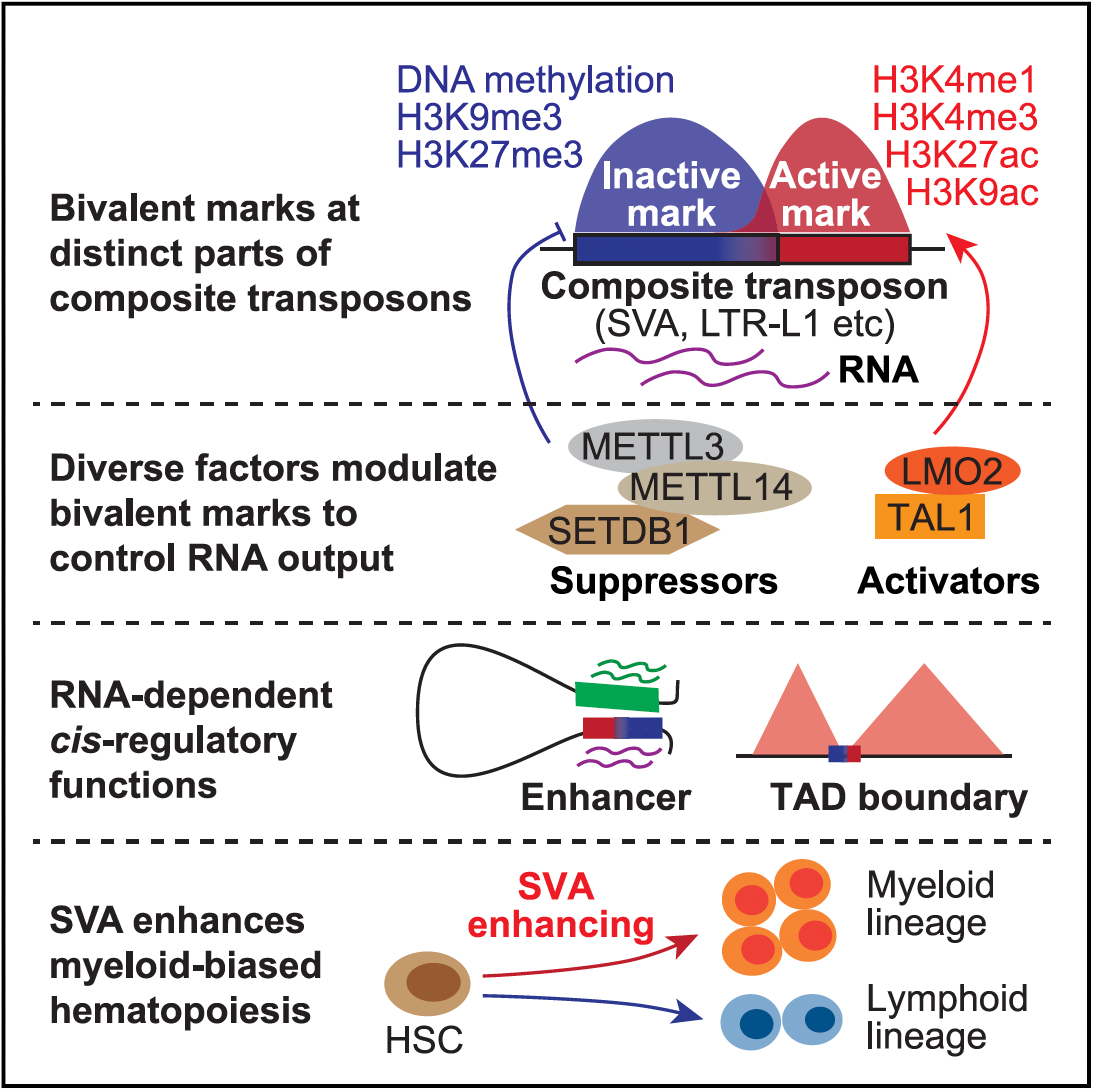
3. Li X, Liu N*. Advances in understanding LINE-1 regulation and function in the human genome. Trends in Genetics (2025). ISSN 0168-9525. [Cover article & Featured Article]

4. Li X#, Bie L#, Wang Y#, Hong Y#, Zhou Z, Fan Y, Yan X, Tao Y, Huang C, Zhang Y, Sun X, Li J, Zhang J, Chang Z, Xi Q, Meng A, Shen X, Xie W, Liu N*. LINE-1 transcription activates long-range gene expression. Nature Genetics 56, 1494–1502 (2024).
News & Views by: Liang, Y., Wang, T. LINE1 mediates long-range DNA interactions. Nature Genetics 56, 1328–1330 (2024).
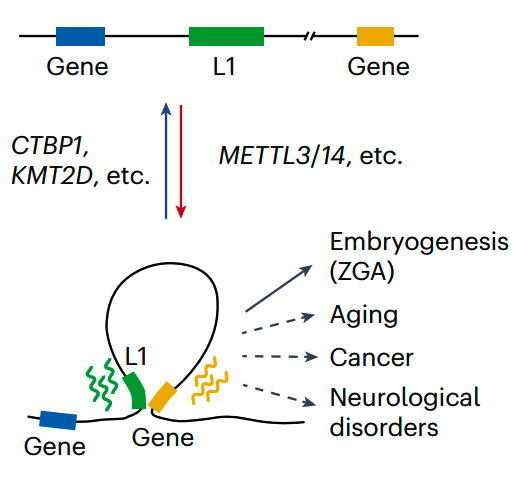
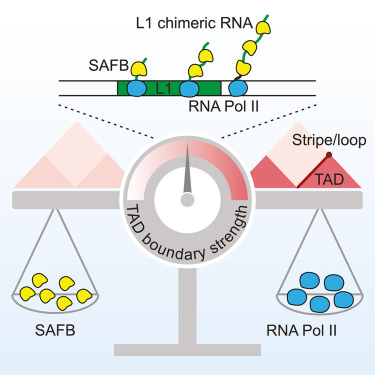
5. Hong Y#, Bie L#, Zhang T#, Yan X, Jin G, Chen Z, Wang Y, Li X, Pei G, Zhang Y, Hong Y, Gong L, Li P, Xie W, Zhu Y, Shen X, Liu N*.SAFB restricts contact domain boundaries associated with L1 chimeric transcription. Molecular Cell (2024). [Featured Article]
6. Wang D, Yan K, Yu H, Li H, Zhou W, Hong Y, Guo S, Wang Y, Xu Chen, Pan C, Tang Y, Liu N, Wu W, Zhang L*, Xi Q*. Fimepinostat impairs NFκB and PI3K/AKT signaling and enhances gemcitabine efficacy in H3K27M-mutated diffuse intrinsic pontine glioma. Cancer Research. (2024) 84(4), p. 598-615
7. Li Y, Yang J, Shen S, Wang W, Liu N, Guo H*, Wei W*. SARS-CoV-2-encoded inhibitors of human LINE-1 retrotransposition. Journal of Medical Virology (2022) DOI: 10.1002/jmv.28135.
8. Zuo F#, Jiang J#, Fu H#, Yan K, Liefke R, Zhang J, Hong Y, Chang Z, Liu N, Wang Z*, Xi Q*. A TRIM66/DAX1/Dux axis suppresses the totipotent 2-cell-like state in murine embryonic stem cells. Cell Stem Cell (2022) 29 (6), 948-961.
(#, co-first author; *, corresponding author)
2018 and Earlier (representative)
9. Liu, N.*, Lee, C.*, Swigut, T., Grow, E., Gu, B., Bassik, M., and Wysocka, J. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 553, 228-232 (2018). [pdf]
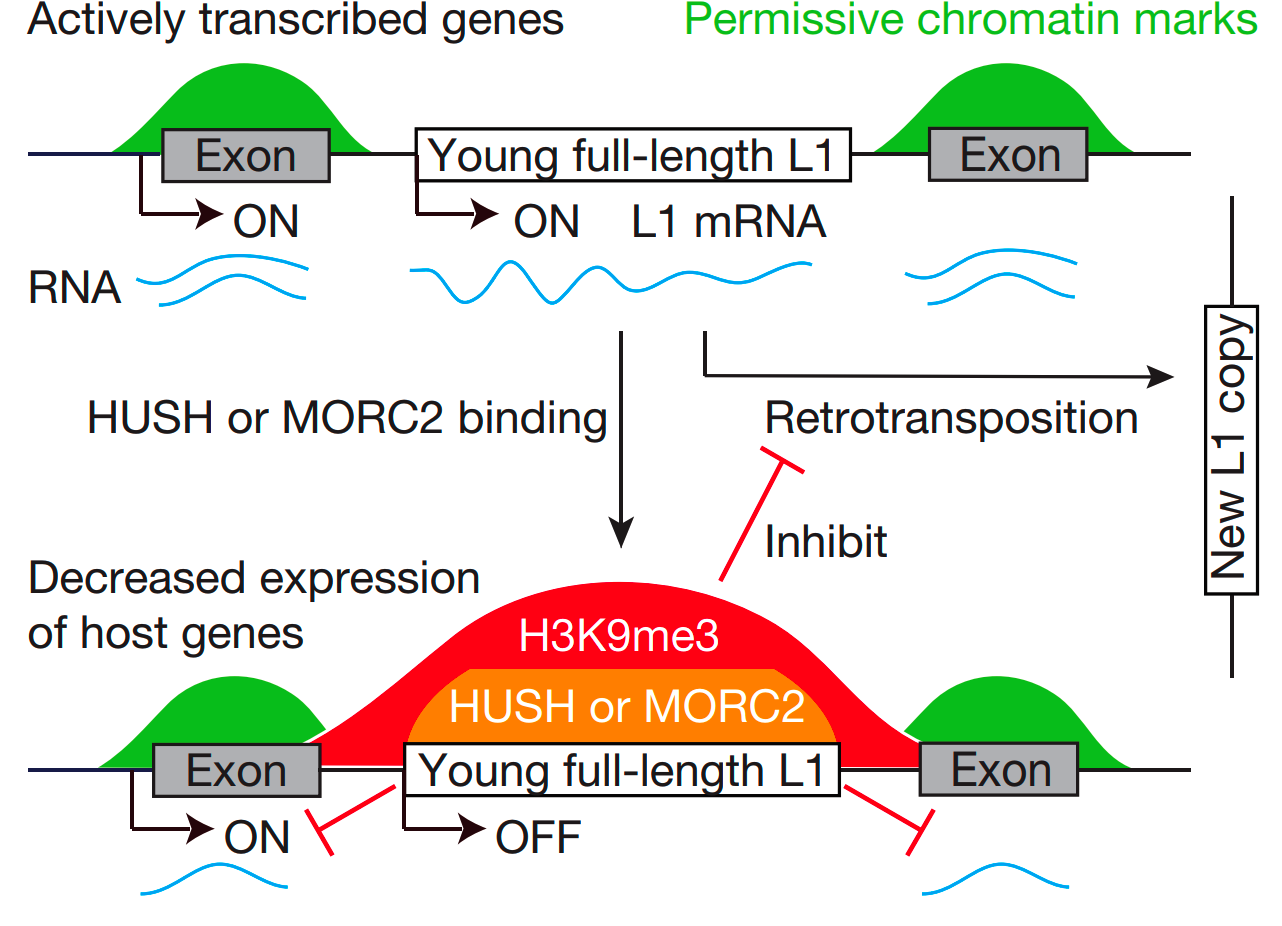
10.Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518, 560-564 (2015). [pdf]
News & Views by: Theler, D., Allain, FT. RNA modification does a regulatory two-step. Nature 518, 492–493 (2015).
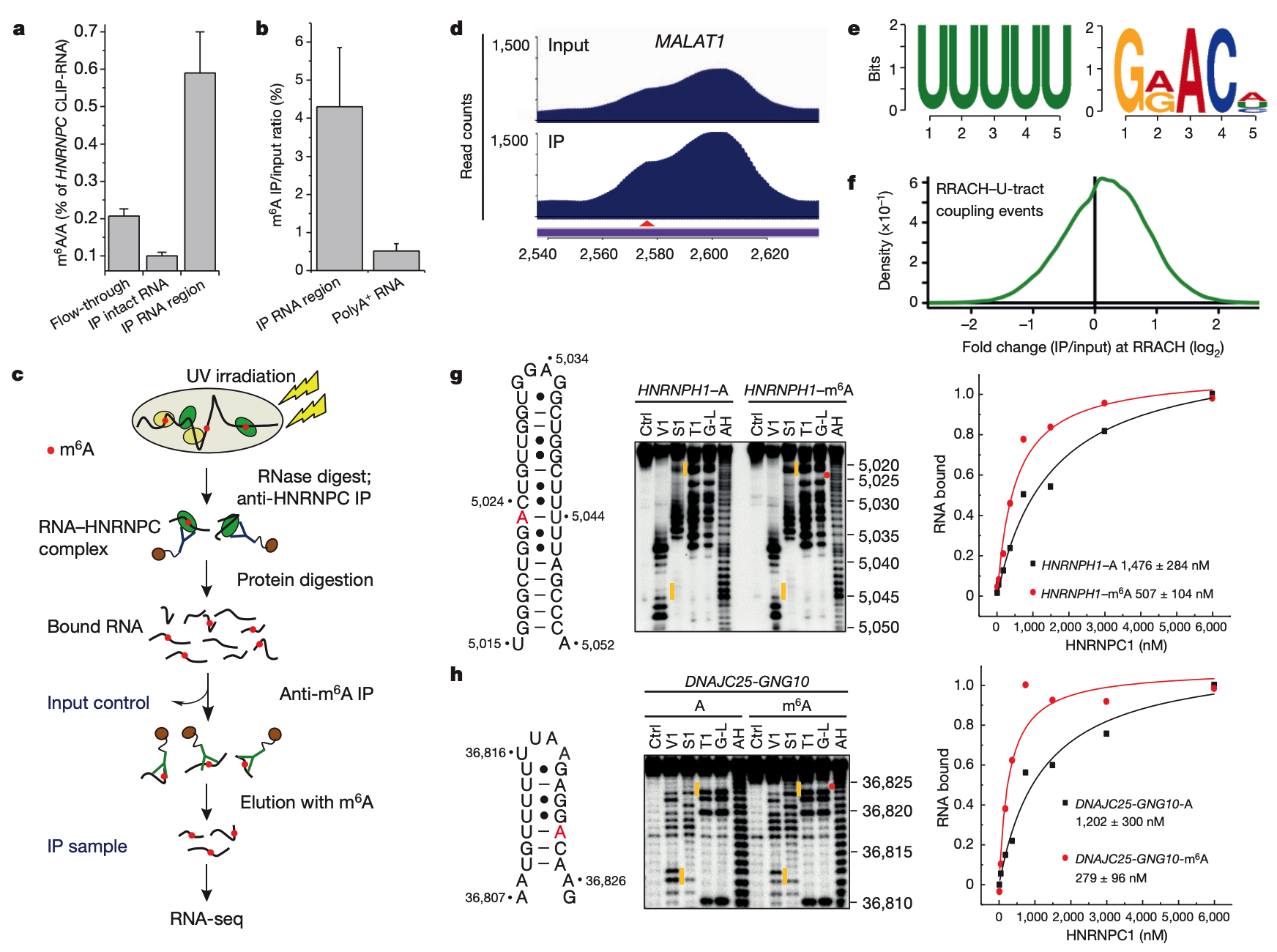
11.Liu, N.*, Pan, T.* N6-methyladenosine-coded RNA Epigenetics. Nat. Struct. Mol. Biol. 23, 98-102 (2016). [pdf]
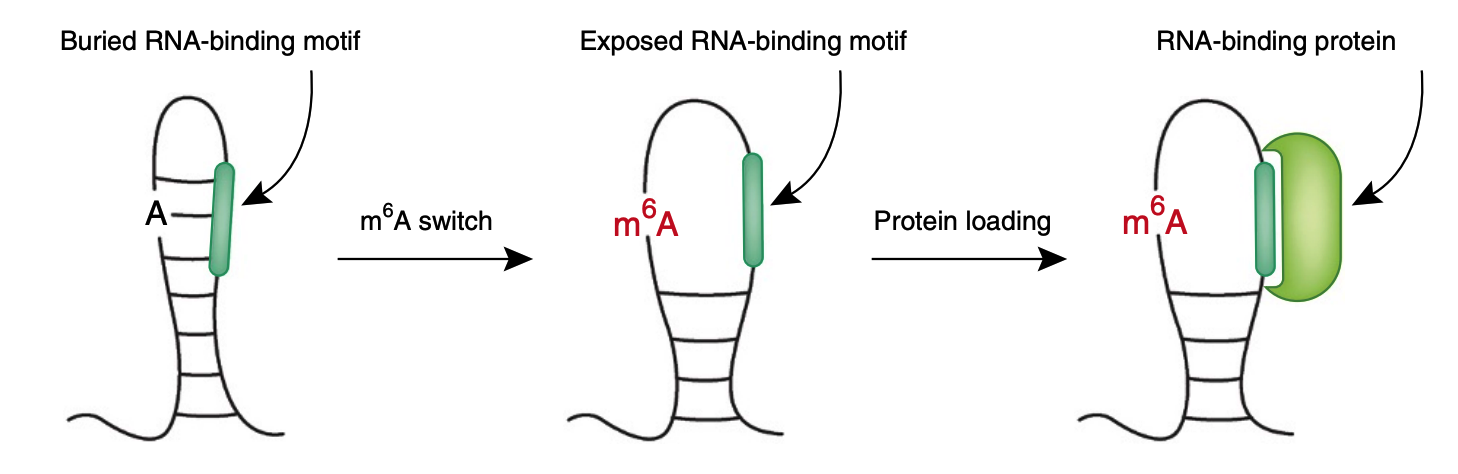
12.Liu, N.*, Zhou, K.*, Parisien, M., Dai, Q., Diatchenko, L., and Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45 (10), 6051-6063 (2017). [pdf]
13.Liu, N., Parisien, M., Dai, Q., Zheng, G., He, C., and Pan, T. Probing N6-methyl-adenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848-1856 (2013). [pdf]
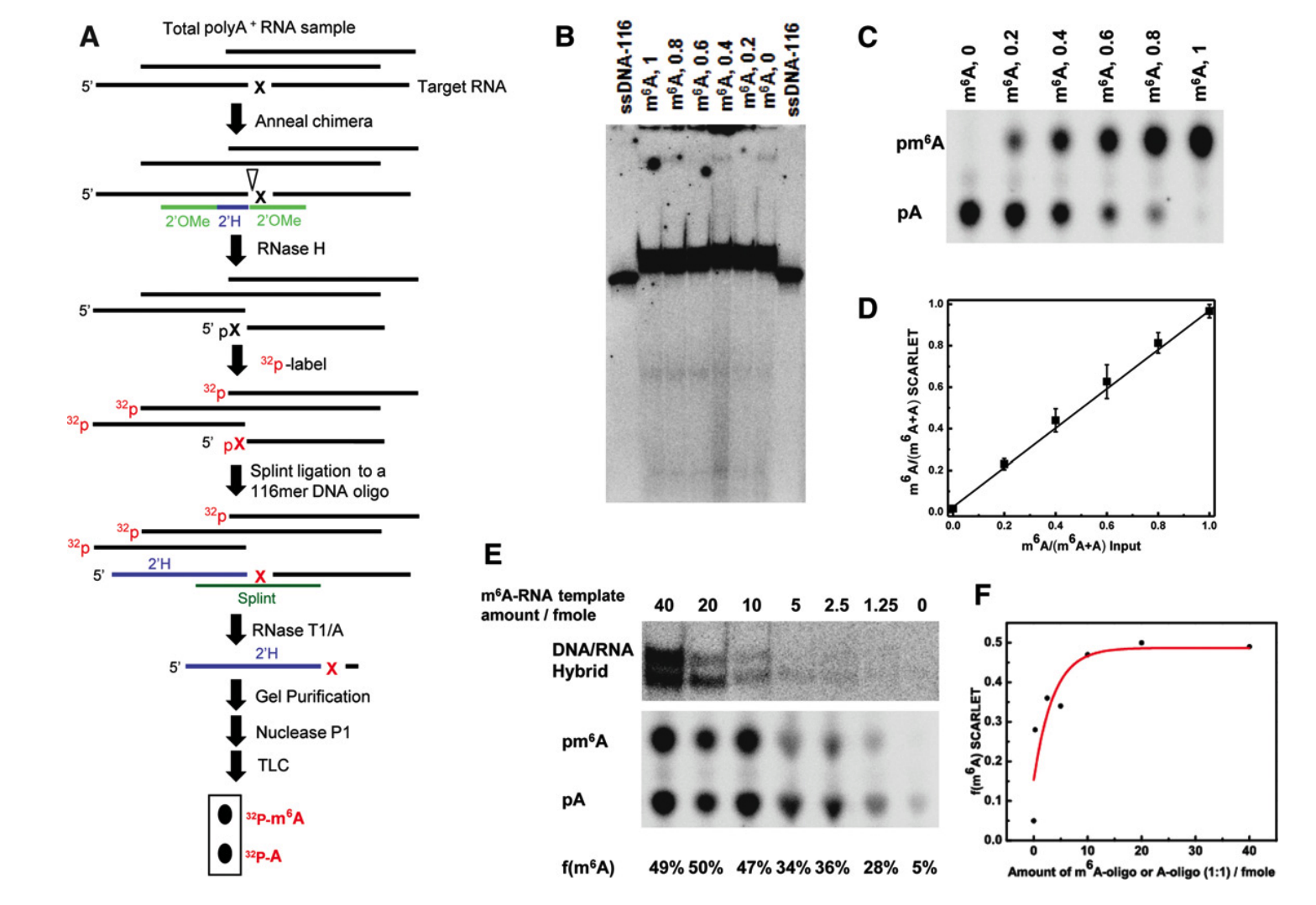
14. Liu, N., Pan, T. RNA epigenetics (Epitranscriptomics). Translating epigenetics to the clinic 19-30 (2017). ( Book chapter)
15. Liu, N.,Pan, T. Probing N6-methyl-adenosine (m6A) RNA modification in total RNA with SCARLET. Post-transcriptional gene regulation 1358, 285-292 (2016). [pdf]
16.Liu, N.,Pan, T. Probing RNA modification status at single nucleotide resolution in messenger and long non-coding RNA. Methods in enzymology 560, 149-159 (2015). ( Book chapter)
17. Liu, N., Pan, T. RNA epigenetics. Translational research 165, 28-35 (2015).[pdf]
18. Zhou, K., Parisien, M., Dai, Q., Liu, N., Diatchenko, L., Sachleben, J., Pan, T. N6-methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. Journal of molecular biology 428, 822-33 (2016). [pdf]
19. Chen, K., Lu, Z., Wang, X., Fu, Y., Luo, G., Liu, N., Han, D., Dominissini, D., Dai, Q., Pan, T., He, C. High-resolution N6-methyladenosine (m6A) map using photo-crosslinking-assisted m6A sequencing. Angewandte chemie 127, 1607-1610 (2015). [pdf]